Abstract
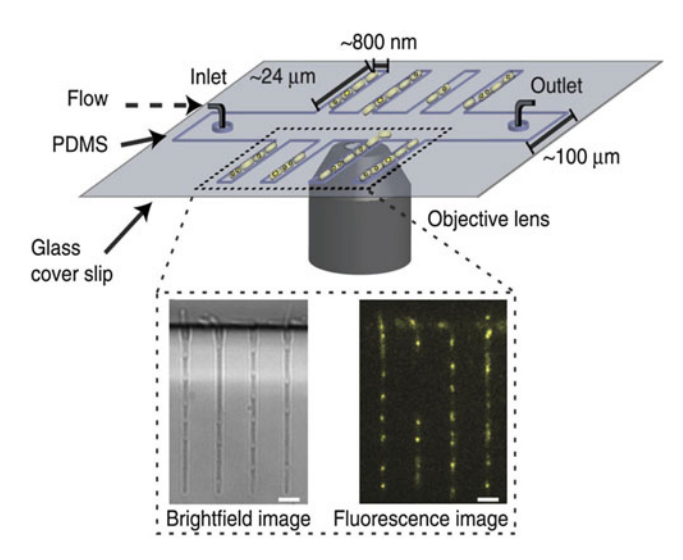
Studying the dynamics of intracellular processes and investigating the interaction of individual macromolecules in live cells is one of the main objectives of cell biology. These macromolecules move, assemble, disassemble, and reorganize themselves in distinct manners under specific physiological conditions throughout the cell cycle. Therefore, in vivo experimental methods that enable the study of individual molecules inside cells at controlled culturing conditions have proved to be powerful tools to obtain insights into the molecular roles of these macromolecules and how their individual behavior influence cell physiology. The importance of controlled experimental conditions is enhanced when the investigated phenomenon covers long time periods, or perhaps multiple cell cycles. An example is the detection and quantification of proteins during bacterial DNA replication. Wide-field microscopy combined with microfluidics is a suitable technique for this. During fluorescence experiments, microfluidics offer well-defined cellular orientation and immobilization, flow and medium interchangeability, and high-throughput long-term experimentation of cells. Here we present a protocol for the combined use of wide-field microscopy and microfluidics for the study of proteins of the Escherichia coli DNA replication process. We discuss the preparation and application of a microfluidic device, data acquisition steps, and image analysis procedures to determine the stoichiometry and dynamics of a replisome component throughout the cell cycle of live bacterial cells.